MOGRIFY® PLATFORM
Systematically predict the transcriptomic switches required
to produce any target cell type from any source cell type.
The MOGRIFY® technology was developed as a systematic means of identifying the optimal combination of transcription factors required to drive cell identity. The platform can be used to enhance existing stem-cell forward programming methods, or bypass development pathways altogether, affecting a direct transdifferentiation between a mature cell type to another mature cell type.
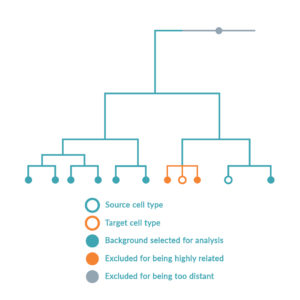
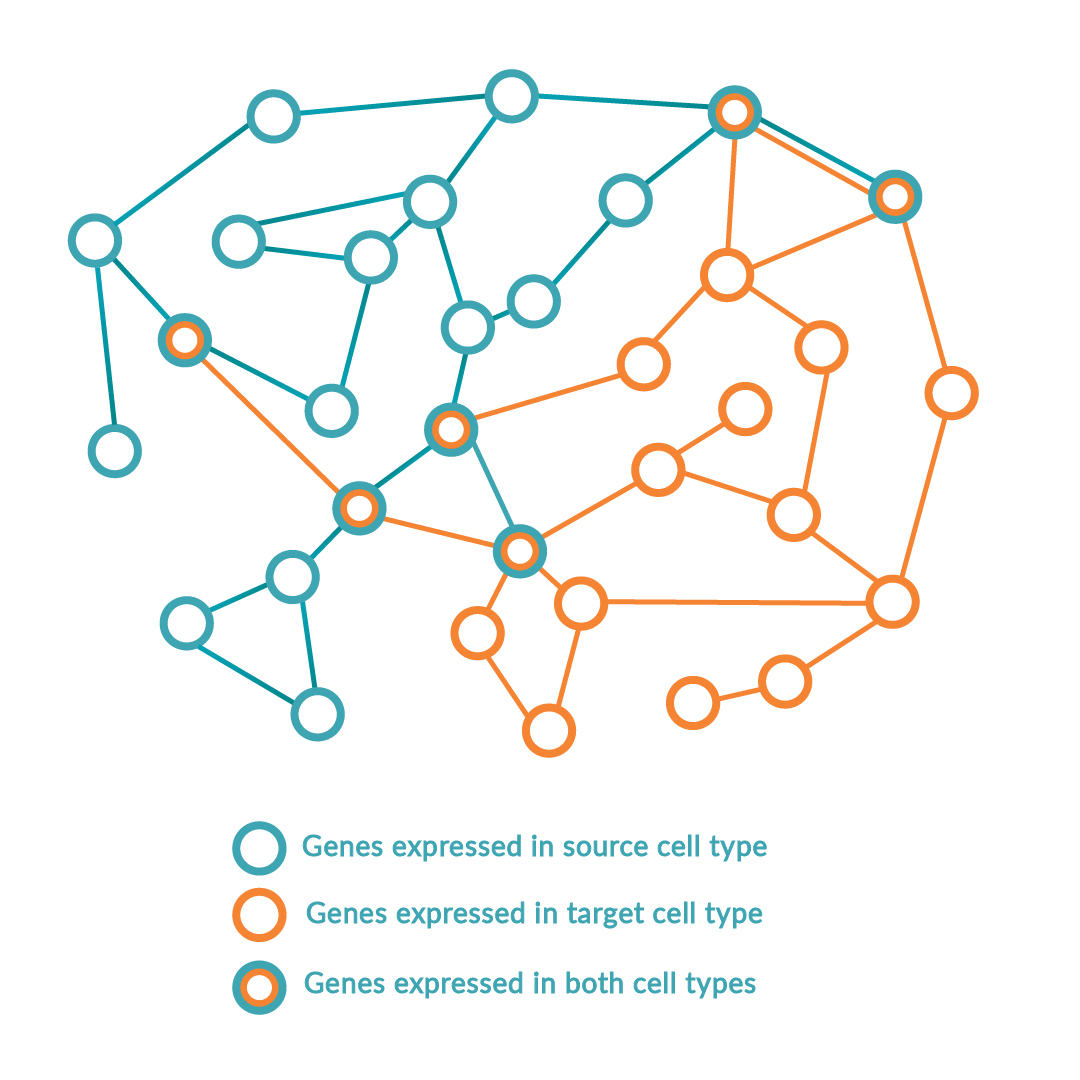
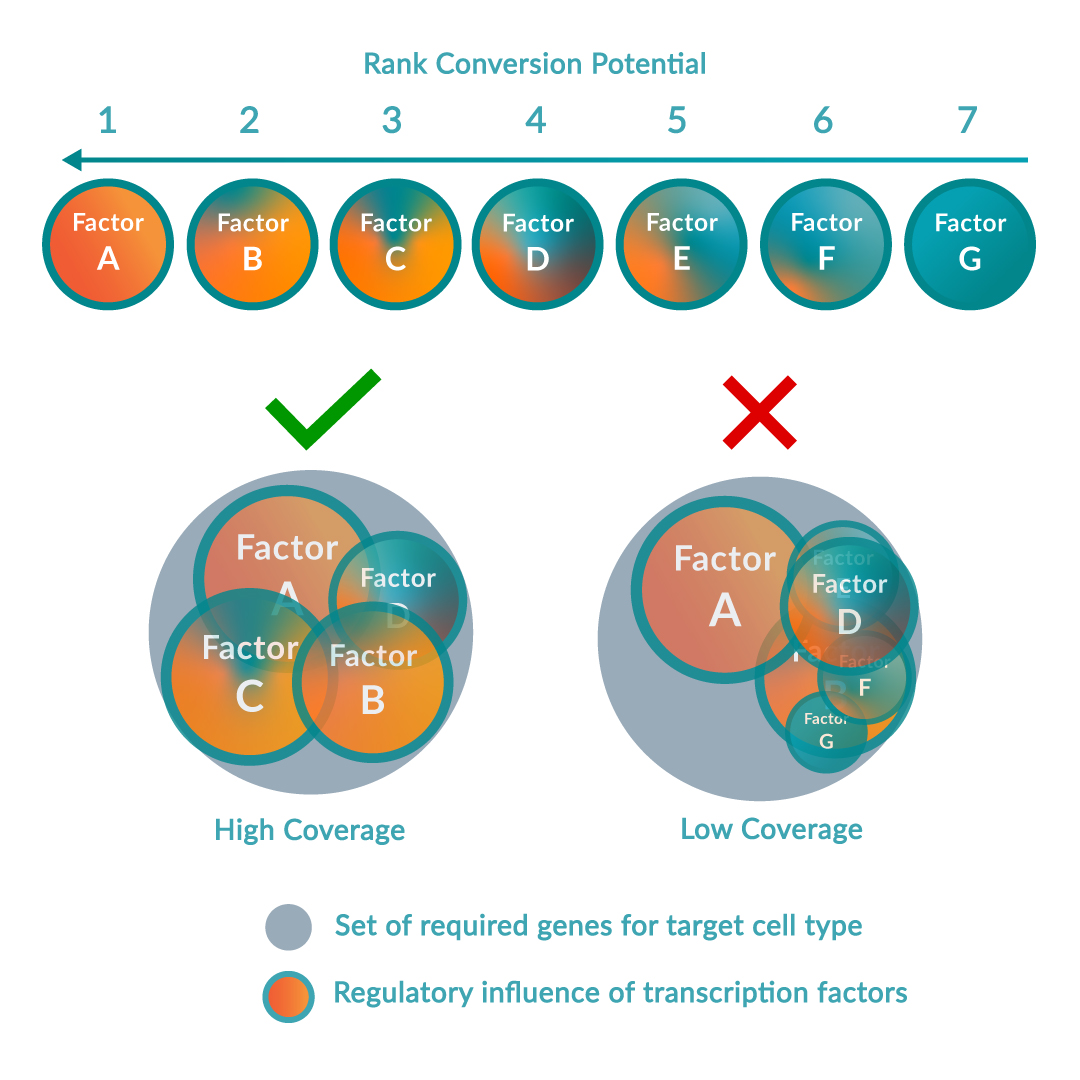
The platform follows a step-wise scientific approach to driving cell identity. Firstly, the RNA of the source and target cell types is sequenced to compare the gene expression levels in the context of all other possible cell types. Simultaneously, local transcriptomic regulatory networks are built around each regulatory gene in the human genome to calculate and rank the effect of the genes on the desired cell conversion by querying large-scale regulatory networks. The optimal combination of key regulators (or transcription factors) is predicted to maximize network coverage while avoiding redundancies. Once this optimal combination has been identified, the DNA sequence of each regulatory gene is encoded into delivery vectors. These are then transduced into the source cell type causing changes in DNA expression profile and consequently switching the genetic programs of the cell, inducing the conversion between source and target cell type.
Over 150 cell types, of which 32 cell conversions (13 successfully validated in vitro) are described in the foundational patent. Images adapted from Rackham OJL et al. A predictive computational framework for direct reprogramming between human cell types. Nature Genetics (2016).