epiMOGRIFY® PLATFORM
Systematically predict the epigenetic switches required
to drive and maintain cell identity.
epiMOGRIFY® is an extension of the Company’s proprietary direct cellular reprogramming technology, MOGRIFY®, that enables the identification of the optimal culture conditions required to maintain cells and support reprogramming in chemically defined media. This can be applied in cGMP manufacture and enhances directed differentiation or transdifferentiation to support the development of scalable off-the-shelf therapies.
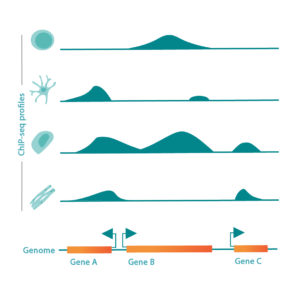
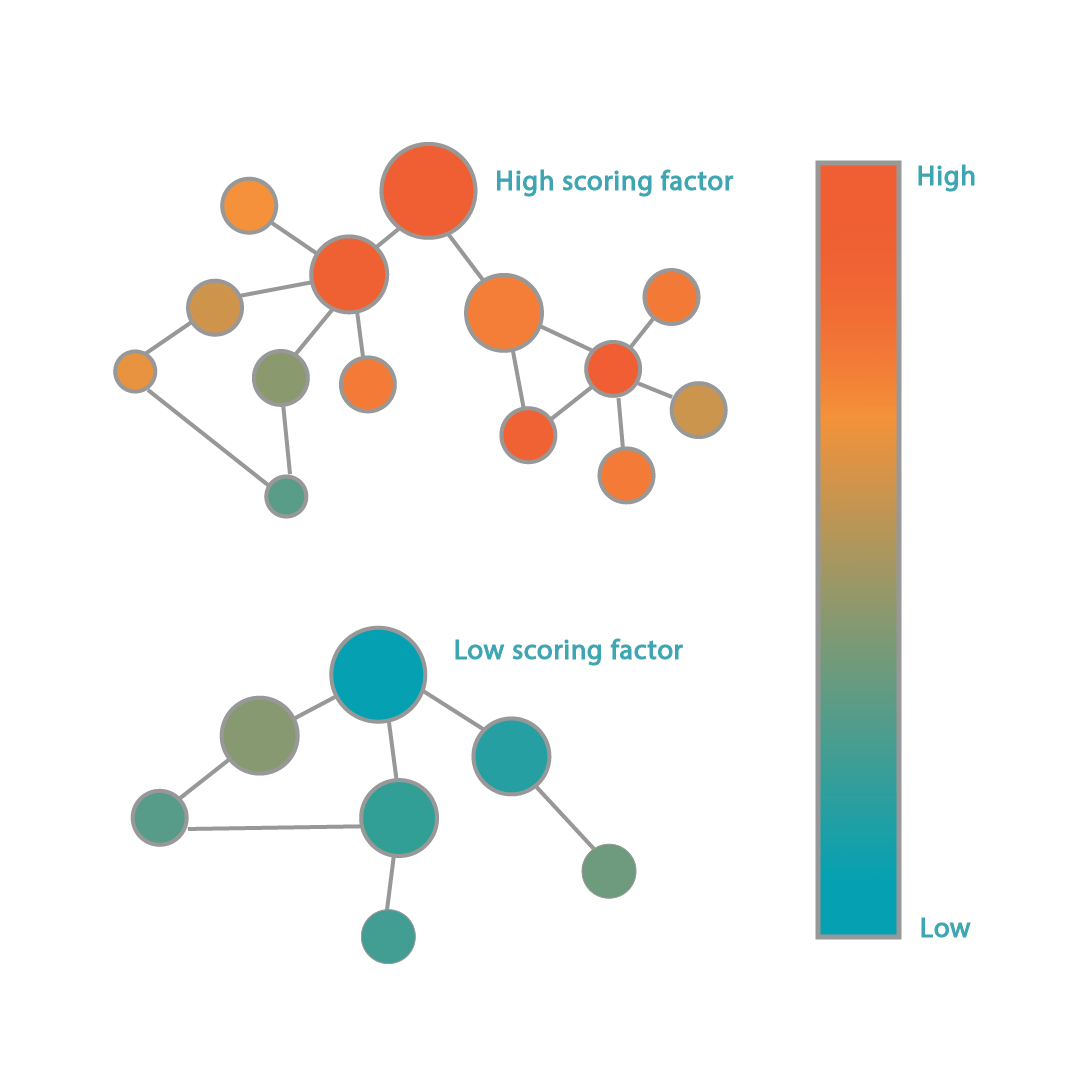
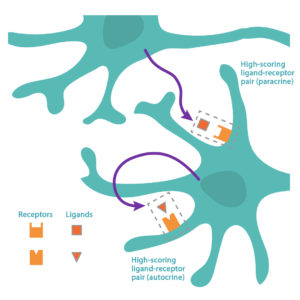
epiMOGRIFY combines gene-regulatory information with a model of a cell’s epigenetic landscape and leverages changes in the level of DNA-histone methylation (H3K4me3 modifications). The platform utilizes data from more than 100 human cell/tissue types to accurately define culture conditions that can maintain the cell identity or induce cell conversion.
The predictive power of epiMOGRIFY has been validated in two ways: cell maintenance and differentiation. epiMOGRIFY-predicted factors are able to maintain astrocytes and cardiomyocytes in vitro in chemically defined media, and promote the generation of astrocytes and cardiomyocytes from neural progenitors and embryonic stem cells, respectively. In both cell maintenance and differentiation, epiMOGRIFY-defined conditions performed as well or better in all cases when compared to existing undefined conditions, significantly increasing cell growth and survival, as well as resulting in a higher differentiation efficiency.
Patent pending on the epiMOGRIFY platform, validated conversion and maintenance of specific cell types. Images adapted from Kamaraj et al., EpiMogrify Models H3K4me3 Data to Identify Signaling Molecules that Improve Cell Fate Control and Maintenance. Cell Systems (2020).